Dweet Mottle Disease
(Citrus Leaf Blotch Virus)
HISTORY, DISTRIBUTION AND IMPORTANCE
In 1968, routine indexing carried out in the Citrus Variety Improvement Program developed in California, revealed a new graft-transmissible disease in a Cleopatra mandarin tree from Florida (CRC 270). Graft inoculation from this source tree to indicator plants induced chlorotic blotching in Dweet tangor and stem pitting in Etrog citron (Roistacher & Blue, 1968). The parent tree showed no visible evidence of damage due to any known virus, the trunk appeared normal and no pitting or bark discoloration was evident, although the fruits were small, the twigs were dying back, and little new growth was apparent. The pathogen responsible of these symptoms was graft and mechanically inoculated to several citrus species and varieties, but symptoms were observed only in Dweet tangor. Therefore it was named Dweet mottle virus (Roistacher & Blue, 1968).
In 1984, in routine indexing done at the Citrus Variety Improvement Program developed in Spain, a graft-transmissible disease was found in Nagami kumquat, clone SRA-153, from Corsica (France). The pathogen present in this kumquat clone induced chlorotic blotches in Dweet tangor (Dm. 01) and stem pitting in Etrog citron (Dm. 02) resembling those of Dweet mottle virus, but additionally it caused vein clearing in Pineapple sweet orange (Dm. 03) and other cultivars, and bud union crease when propagated on Troyer citrange rootstock (Dm. 04) (see bud union disorders in Nagami kumquat on trifoliate rootstocks) (Navarro et al., 1984; Galipienso et al., 2000). Citrus leaf blotch virus (CLBV) was partially purified from Nagami kumquat SRA-153 (Galipienso et al., 2001) and its genomic RNA (gRNA) completely sequenced (Vives et al., 2001). Later, CLBV was detected by RT-PCR in two sources of Dweet mottle from California (DMV-931 and DMV-932) using primers directed to the replicase and coat protein genes. The nucleotide identity between CLBV isolate SRA-153 and Dweet mottle sources DMV-931 and DMV-932 was 96.6 and 96.8% in the replicase, and 98.7 and 98.5% in the coat protein gene, respectively. These data and previous finding that Dweet mottle and CLBV are difficult to eliminate by shoot-tip grafting or thermotherapy, suggest that Dweet mottle may be caused by CLBV (Vives et al.,2006).
An infectious full-genome cDNA clone (CLBV-IC) was obtained from CLBV isolate SRA-153. CLBV-IC only induced chlorotic blotching in Dweet tangor plants and stem pitting in Etrog citron plants, but no vein clearing in Pineapple sweet orange or bud union crease on trifoliate rootstocks. These results suggest that bud union crease and vein clearing symptoms are likely caused by a different pathogen or by an interaction between CLBV and other biotic factors.
CLBV was detected in three mandarins imported from Japan through the National Citrus Quarantine Station, in freeze-dried tissue of a Nagami kumquat from New South Wales (Australia) (Dm. 05), in several trees of Roble sweet orange grafted on Swingle citrumelo (Dm. 06) from commercial orchards in Florida (USA), and in several trees grafted on citrange rootstock from Valencia (Spain) showing decline and bud union crease (Dm. 07) (Galipienso et al., 2001, 2004; Vives et al., 2002a), although it was not detected in other trees with similar symptoms, sometimes growing in their close vicinity. Low genetic variation was found between CLBV isolates from different host species and geographical origins, indicating genetic stability of CLBV (Vives et al., 2002b).
The economic importance of CLBV for the citrus industry is presently unknown since its incidence in different citrus areas has not been evaluated and the actual involvement of CLBV with other pathogen in the bud union crease has not yet been proved.
NAME OF DISEASE AND SYNONYMS
Dweet mottle (Roistacher & Blue, 1968) or Citrus Leaf blotch (Galipienso et al., 2001; Vives et al., 2006).
Abbreviation: Dm for Dweet mottle.
SYMPTOMATOLOGY
General aspect of affected field tree
CLBV has been detected only in trees showing bud union crease on trifoliate rootstocks, but detection failed in other trees with similar symptoms growing nearby. Involvement of CLBV in bud union crease has not been demonstrated.
Symptoms on trunk
Stem pitting on Etrog citron resembles that induced by Citrus tristeza virus in the same indicator plant (Dm. 02).
Symptoms on leaves
Chlorotic botching in Dweet tangor (Dm. 01). This symptom could be confused with those produced by other diseases like Psorosis, Cristacortis, Impietratura and Concave Gum in the same indicator plant.
Symptoms on fruit
None.
Histological and cytological symptoms
No histological and cytological symptoms reported.
CAUSAL AGENT: DESCRIPTION AND PROPERTIES
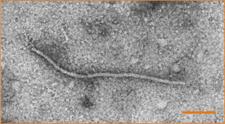
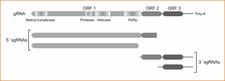
CLBV is a member of the Flexiviridae family. CLBV virions are filamentous particles about 960 ´ 14 nm in size (Dm. 08), containing a single-stranded, positive sense, genomic RNA (gRNA), encapsidated by a 42-kDa coat protein (Galipienso et al., 2001). The gRNA has 8747 nt organized in three open reading frames (ORFs) and untranslated regions of 73 and 541 nt at the 5´ and 3´ termini, respectively. ORF1 potentially encodes a large polyprotein (227,4 kDa) containing methyltransferase, papain-like protease, helicase and RNA-dependent RNA polymerase (RdRp) motifs; ORF2 encodes a 40,2 kDa polypeptide containing a motif characteristic of the 30K superfamily of cell-to-cell movement proteins; and the 40,7 kDa polypeptide encoded by ORF3 was identified as the coat protein. Biological and molecular differences with other known viruses support the idea of including it in a new genus, for which the name citrivirus was proposed (Vives et al., 2001). In addition to the gRNA, CLBV infected tissues contain two subgenomic RNAs (sgRNAs) 3´ coterminal with the gRNA and two that are 5´ coterminal (Vives et al., 2002c) (Dm. 09).
HOST RANGE
Many Citrus ssp(sweet orange, sour orange, grapefruit, mandarin and lemon), and species in other genera of the family Rutaceae like Poncirus trifoliata and its hybrids citrange and citrumelo can host CLBV.
The virus has been experimentally transmitted from citrus to Nicotiana occidentalis and Nicotiana benthamiana, that show symptomless infections.
Sensitive species, varieties or combinations
Dweet tangor and Etrog citron.
Symptomless species, varieties or combinations
• Immune: none described
• Tolerant: all citrus cultivars assayed except Dweet tangor and Etrog citron showed symptomless infections.
TRANSMISSION
Natural
Cultural transmission through infected budwood.
Seed transmission has been reported in Carrizo citrange, Nagami kumquat or sour orange (Citrus aurantium L.) at a rate of 2.5% (Guerri et al., 2004).
No evidence for vector transmission.
Experimental
The virus has been experimentally transmitted from citrus to citrus by stem-slash inoculation and from citrus to N. occidentalisand N. benthamiana by rubbing carborundum-dusted leaves with semipurified viral extracts.
EPIDEMIOLOGY
Not studied.
DIAGNOSIS
Diagnostic field symptoms
Non reported
Biological indexing
Since CLBV induced leaf symptoms in Dweet tangor (Dm. 01) and stem pitting in Etrog citron (Dm. 02), these two species are appropriate diagnostic indicators. Chlorotic leaf blotching in Dweet tangor is faster to appear and easier to observe, therefore, inoculation of that indicator is recommended as routine indexing procedure. To reduce the risk of false negatives due to low titer and uneven distribution within infected plants of some citrus cultivars, at least six indicator plants graft-inoculated with four pieces of bark from the candidate tree should be used in each test, and at least two or three flushes should be observed for leaf symptoms. Indicator plants should be decapitated after inoculation and preferably incubated in a temperature controlled glasshouse (26ºC day and 18ºC night). Chlorotic leaf blotching in Dweet tangor usually appears in tender fully expanded leaves and can be observed in successive flushes with similar intensity, but it fades with leaf hardening. The symptoms usually appears in young shoots located above or below the inoculation point, but not in shoots growing at the opposite side of the stem (Galipienso et al., 2000).
Serological and molecular diagnostic methods
Serological methods are not available.
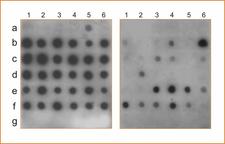
CLBV can be detected by molecular hybridization using digoxigenin-labeled cDNA probes (DIG-probes) and nucleic acid extracts (dot-blot hybridization, DBH) (Dm. 10) or tissue prints (tissue print hybridization, TPH) (Dm. 11), and by one step reverse transcription and polymerase chain reaction (RT-PCR) (Dm. 12) (Vives et al., 2002a; Galipienso et al.,2004). DBH or TPH allow reliable detection of CLBV in tender shoots or young leaves from greenhouse-grown plants, but detection in old hardened leaves was inconsistent and did not allow a reliable diagnostic (Dm. 10, 11). One-step RT-PCR is 10-fold more sensitive than DBH or TPH and amplification can be obtained with all infected tissues (Dm. 12). In Pineapple sweet orange, and perhaps in other varieties not tested, analysis of at least ten individual samples per tree is necessary to reduce the risk of false negatives. These results agree with those obtained by biological indexing (Galipienso et al., 2000).
CLBV detection was less consistent in field trees than in greenhouse-grown plants (Dm. 13), possibly due to lower virus accumulation and/or uneven virus distribution within infected trees.
CONTROL
As CLBV is seed transmitted, exclusion of the virus during citrus propagation requires using, not only virus-free buds, but also rootstock seedlings that originate from CLBV-free seed source trees.
SELECTED REFERENCES
Galipienso, L., L. Navarro, J.F. Ballester-Olmos, J. Pina, P. Moreno, J. Guerri (2000). Host range and symptomatology of a graft-transmissible pathogen causing bud union crease of citrus on trifoliate rootstocks. Plant Pathol. 49: 308-314.
Galipienso, L., M.C. Vives, P. Moreno, R.G. Milne, L. Navarro, J. Guerri (2001). Partial characterisation of Citrus leaf blotch virus, a new virus from Nagami kumquat. Arch. Virol. 146: 357-368.
Galipienso. L., M.C. Vives, L. Navarro, P. Moreno, J. Guerri (2004). Detection of Citrus leaf blotch virus using digoxigenin-labeled cDNA probes and RT-PCR. Eur. J. Plant Pathol. 110: 175-181.
Guerri, J., J.A. Pina, M.C. Vives, L. Navarro, P. Moreno (2004). Seed transmisión of Citrus leaf blotch virus: implications in quarantine and certification programs. Plant Dis. 88: 906.
Navarro, L., J.A. Pina, J.F. Ballester-Olmos, P. Moreno, M. Cambra (1984). A new graft transmissible disease found in Nagami kumquat. p. 234-240. In S.M. Garnsey, L.W. Timmer & J.A. Dodds (eds) Proc. 9th Conf. Intern. Organization Citrus Virol., IOCV, Riverside, CA.
Roistacher, C.N., R.L. Blue (1968). A psorosis-like virus causing symptoms only on Dweet tangor. p. 13-18. In J.F.L. Childs (ed) Proc. 4th Conf. Intern. Organization Citrus Virol.,. Univ. Florida Press, Gainesville.
Vives, M.C., L. Galipienso, L. Navarro, P. Moreno, J. Guerri (2001). The nucleotide sequence and genomic organization of Citrus leaf blotch virus: candidate type species for a new virus genus. Virology 287: 225-233.
Vives, M.C., L. Galipienso, L. Navarro, P. Moreno, J. Guerri (2002a). Citrus leaf blotch virus: a new citrus virus associated with bud union crease on trifoliate rootstocks. p. 205-212. In N. Duran-Vila, R.G. Milne & J.V. da Graça (eds) Proc. 15th Conf. Intern. Organization Citrus Virol., IOCV, Riverside, CA.
Vives, M.C., L. Rubio, L. Galipienso, L. Navarro, P. Moreno, J. Guerri (2002b). Low genetic variation between isolates of Citrus leaf blotch virus from different host species and different geographical origins. J. Gen. Virol. 83: 2587-2591.
Vives, M.C., L. Galipienso, L. Navarro, P. Moreno, J. Guerri (2002c). Characterization of two kinds of subgenomic RNAs produced by Citrus leaf blotch virus. Virology 295: 328-336.
Vives, M.C., J.A. Pina, J. Juárez, L. Navarro, P. Moreno, J. Guerri (2006). Dweet mottle disease probably is caused by Citrus leaf blotch virus. p. 251-256. In N. Duran-Vila, M. Hilf & M. Rocha-Peña (eds) Proc. 16th Conf. Intern. Organization Citrus Virol., IOCV, Riverside, CA.
Prepared (2007) by M.C. Vives, P. Moreno and J. Guerri
Centro de Protección Vegetal y Biotecnología
(I.V.I.A.)
Crta. Moncada-Náquera km 4.5
46113 Moncada
Valencia
(Spain)
Click any image to see it larger.
PHOTOS |
LEGENDS AND AUTHORS |
|---|---|
|
Chlorotic blotching in a Dweet tangor leaf induced by Nagami kumquat SRA-153. (Spain) – L. Galipienso
|
|
|
Stem pitting in Etrog citron induced by Nagami kumquat SRA-153. (Spain) – L. Galipienso
|
|
|
Vein clearing in a Pineapple sweet orange leaf induced by Nagami kumquat SRA-153. (Spain) – L. Galipienso
|
|
|
Bud union crease of a propagation of Nagami kumquat SRA-153 on Troyer citrange rootstock (left) compared with normal union of a healthy control plant (right). (Spain) – L. Galipienso
|
|
|
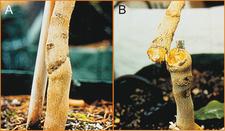
Bud union crease of a propagation of Nagami kumquat infected with CLBV on Troyer citrange rootstock (A). Brittle union of the infected plant (B). (Australia) – P. Barkley
|
|
|
Bud union crease symptoms of a declining Roble sweet orange on citrumelo tree infected with CLBV: wood crack at the bud union line and crests in the inner bark corresponding with the wood crack. (Florida) – P. Moreno
|
|
|
Decline of a 20-year-old Navelina sweet orange on Troyer citrange tree with bud union crease, infected with CLBV. (Spain) – J. Guerri
|
|
|
Electron micrograph of a CLBV virion purified from kumquat SRA-153, stained with 1 % uranyl acetate. Bar = 200 nm. – R. Milne
|
|
|
Genomic organization of CLBV and 5´- and 3´-coterminal subgenomic RNAs. – M.C. Vives
|
|
|
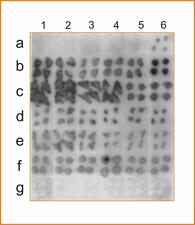
Detection of CLBV by dot blot hybridization using a probe of the RdRp region and total RNA extracts from young leaves (left panel) or old leaves (right panel) of the following citrus varieties: Pineapple sweet orange (a), Eureka lemon (b), Marsh grapefruit (c), Nules clementine (d), Navelina sweet orange (e) and Nagami kumquat infected with CLBV (f) or healthy (g). Numbers on the top correspond to different samples of the same plant (1–6). Nagami kumquat infected with CLBV isolate SRA-153 was used to graft inoculate the other citrus varieties. – M.C. Vives
|
|
|
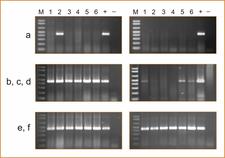
Tissue print hybridization of the RdRp probe with imprints of young shoots from Pineapple sweet orange (a), Eureka lemon (b), Marsh grapefruit (c), Nules clementine (d), Navelina sweet orange (e) and Nagami kumquat infected with CLBV isolate SRA-153 (f) or healthy (g). Numbers on the top (1–6) correspond to different shoots. Nagami kumquat infected with CLBV isolate SRA-153 was used to graft inoculate the other citrus varieties. – M.C. Vives
|
|
|
Detection of CLBV by RT–PCR with primers of RdRp region using total RNA extracts from young leaves (left panels) or old leaves (right panels) of the following citrus varieties: Pineapple sweet orange (a), Eureka lemon (b), Marsh grapefruit (c), Nules clementine (d), Navelina sweet orange (e) and Nagami kumquat infected with CLBV (f) or healthy (g). Numbers on the top correspond to different samples of the same plant (1–6). Nagami kumquat infected with CLBV isolate SRA-153 was used to graft inoculate the other citrus varieties, and was also included as positive (+) control for RT–PCR. Healthy kumquat was used as negative (−) control. – M.C. Vives
|
|
|
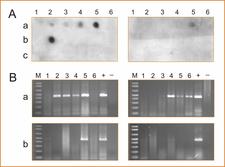
Detection of CLBV by dot blot hybridization (A) or RT–PCR (B) using total RNA extracts from young leaves (left panels) and old leaves (right panels) of two CLBV-infected Navelina sweet orange field trees (a, b) and a healthy greenhouse-grown Navelina plant (c). Numbers on the top (1–6) correspond to different samples of each plant. Nagami kumquat infected with CLBV isolate SRA-153 (+) and healthy kumquat (−) were used as positive and negative controls for RT–PCR. The probe and primers of RdRp region were used for DBH and RT–PCR, respectively.– M.C. Vives
|